Your All-In-One Bioinformatics Software Solution

Over 15k researchers choose OmicsBox for their projects!
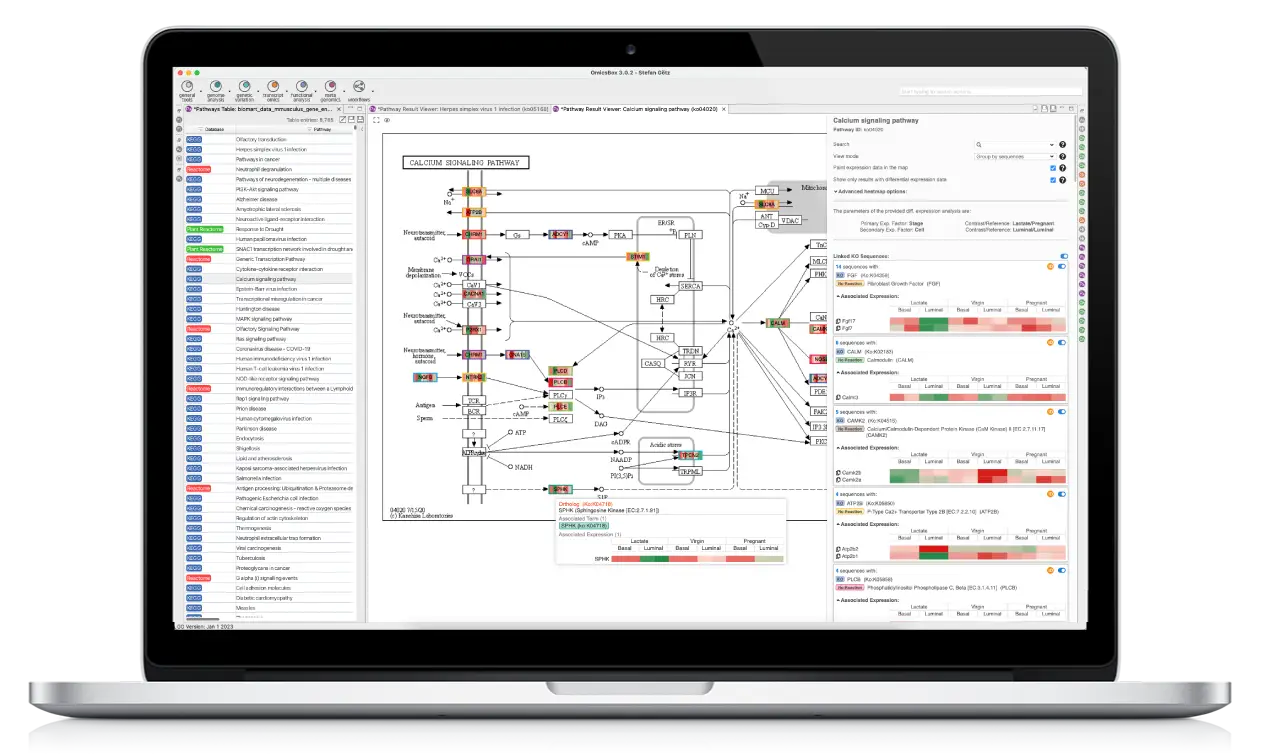
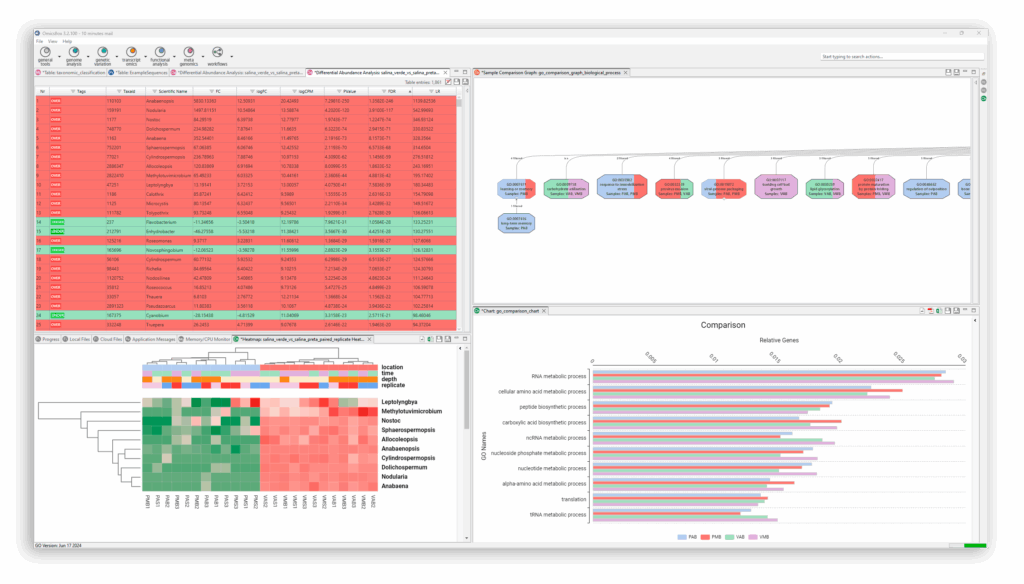
OmicsBox is the leading bioinformatics solution that offers end-to-end NGS data analysis of genomes, transcriptomes, and metagenomes.
We have designed OmicsBox to be user-friendly, efficient, and with a powerful set of tools to extract biological insights from omics data.
OmicsBox is used by top private and public research institutions worldwide. It allows researchers to easily process large and complex data sets, and streamline their analysis process.
Five modules to easily process large and complex data sets
From raw reads to functional insights fast and easy
Identify and analyze genetic variations within a population or a species.
Allows different types of microbiome analysis like assembly and taxonomic classification.
Easily convert raw DNA-Seq reads into a structurally annotated and curated draft genome.
Bioinformatics, simplified. Focus on what matters.
Designed for Scientists: Analyze. Visualize. Interpret.
- No coding or programming skills needed
- Easy to use: end-to-end, modular design
- Process large datasets with cloud support vía OmicsCloud
- Flexible licensing for labs and institutions
- Features always up-to-date: free updates and improvements
- Dedicated support team: 48h response for issues
- Management Dashboard for susbscription and users
Over 900 Institutions around the world trust OmicsBox










Take your research to the next level with OmicsBox
Get to Know Us

BioBam is a leading bioinformatics solution provider which accelerates research in disciplines such as agricultural genomics, microbiology, and environmental NGS studies, amongst others.
At BioBam we are committed to the development of user-friendly software solutions for biological research. Our mission is to transform complex data analysis procedures into attractive and interactive tasks. Our ultimate goal is to close the gap between experimental work, bioinformatics analysis, and applied research.
Latest Blog Articles


Join & Call vs. Call & Join: Poster at ISMB/ECCB2025
Fabian Jetzinger
August 4, 2025

BioBam at the LongTREC Summer School
Fabian Jetzinger
July 18, 2025

Differences between Fisher’s Exact Test and GSEA for Functional Enrichment
Marta Benegas Coll
July 8, 2025

Reference-free analysis of long-read RNA sequencing for non-model species
Fabian Jetzinger
February 3, 2025