How to create a gene list within OmicsBox to run the functional enrichment analysis
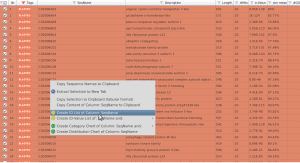
This article explains how to create ID lists of either Blast results or differential expressed genes in Blast2GO.

This article explains how to create ID lists of either Blast results or differential expressed genes in Blast2GO.
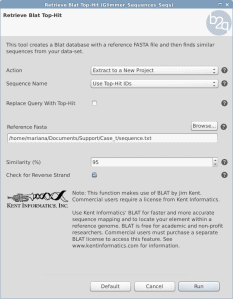
When running GeneFinding the sequences receive a name with the predicted genes. The first part of the sequence identifier comes from the genome reference sequence name (de-novo assembly) and then a _orfx is appended, where x is a number. Sometimes this name is not useful to proceed with downstream analysis or compare results from other experiments. Is there any way
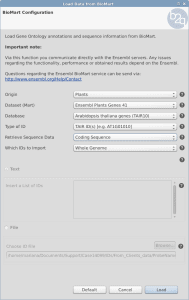
OmicsBox/Blast2GO offers two different features to retrieve the gene/protein sequences as well as the corresponding annotation from a list of identifiers within Blast2GO PRO. Both features can be found under File > Load > Load Annotations. The expected input file is a text file with the identifiers in a single column without a header.
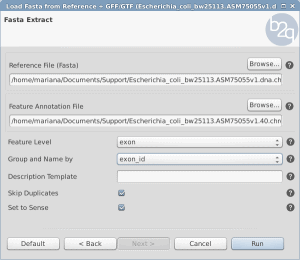
Sometimes databases provide the whole genome and the GFF or GTF files but not the exon or CDS FASTA files.With OmicsBox/Blast2GO it is possible to load a Fasta sequences and to extract the exons or the CDS from the genome using the GFF file.
OmicsBox allows combining two Functional Anlaysis projects which contain either have the same identifiers or different ones.

Blast2GO Supported Project. Researchers: Mrs.Soulmaz Ekhtari Teachers: Dr. Jafar Razeghi, Dr. Karim Hasanpur, and Dr. Arash Kianianmomeni Background and Project Overview: The evolutionary origin of some multicellular organisms has not yet been completely studied. A clear characteristic of complex multicellular organisms is germ-soma differentiation. The monophyletic group of volvocine contains multiple algae species ranging from unicellular species such as Chlamydomonas reinhardtii

(Release Date: 02/08/2018) We are happy to announce that Blast2GO 5.2 has now been released. This update makes our bioinformatics tool for straightforward functional genomics even better.Read below to learn which features and improvements are part of this release. Main features: Venn Diagrams that are fully customizable and allows multiple list comparisons FastQ Pre-Processing (Adapters, Quality, Length) with multiple trimming options.
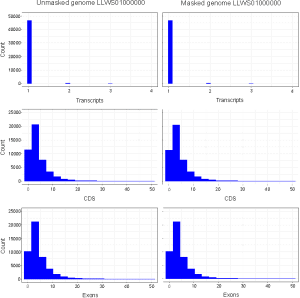
Does RNA-seq-based Eukaryotic GeneFinding of Blast2GO require repeat-masking the whole genome shotgun (WGS) sequence? A case study in jute (Corchorus olitorius L., Malvaceae s. l.) Debabrata Sarkar1, Carlos Menor2 and Nagendra Kumar Singh3 1Biotechnology Unit, Division of Crop Improvement, ICAR-Central Research Institute for Jute and Allied Fibres (CRIJAF), Nilganj, Barrackpore, Kolkata 700120, West Bengal, India. E-mail: debabrata.sarkar@icar.gov.in. ORCID iD: 0000-0003-3943-96462Blast2GO Team,
Blast2GO Supported Project. Researchers: Prof. Luis Delaye, CINVESTAV Irapuato, México Prof. Enrique Calderon, Instituto de Biomedicina de Sevilla, España Prof. Andrés Moya, I2SysBio, Universidad de Valencia, España Background and Project Overview: Fungi from the genus Pneumocystis parasites lungs of mammals. These fungi show extensive stenoxenism, meaning that each Pneumocystis specie parasites a single mammal species. In humans, Pneumocystis causes pneumonia
