Our Teammate and PhD student Fabian Jetzinger, part of the LongTREC MSCA doctoral network, recently spent two months in Stockholm to collaborate with the developers of the isONpipeline tools. This pipeline, part of the OmicsBox Transcriptomics module, facilitates reference-free transcriptome reconstruction from Oxford Nanopore or PacBio long-read RNA sequencing data. Learning from their expertise and bringing in his own set of fresh ideas, Fabian managed to implement improvements to the software.
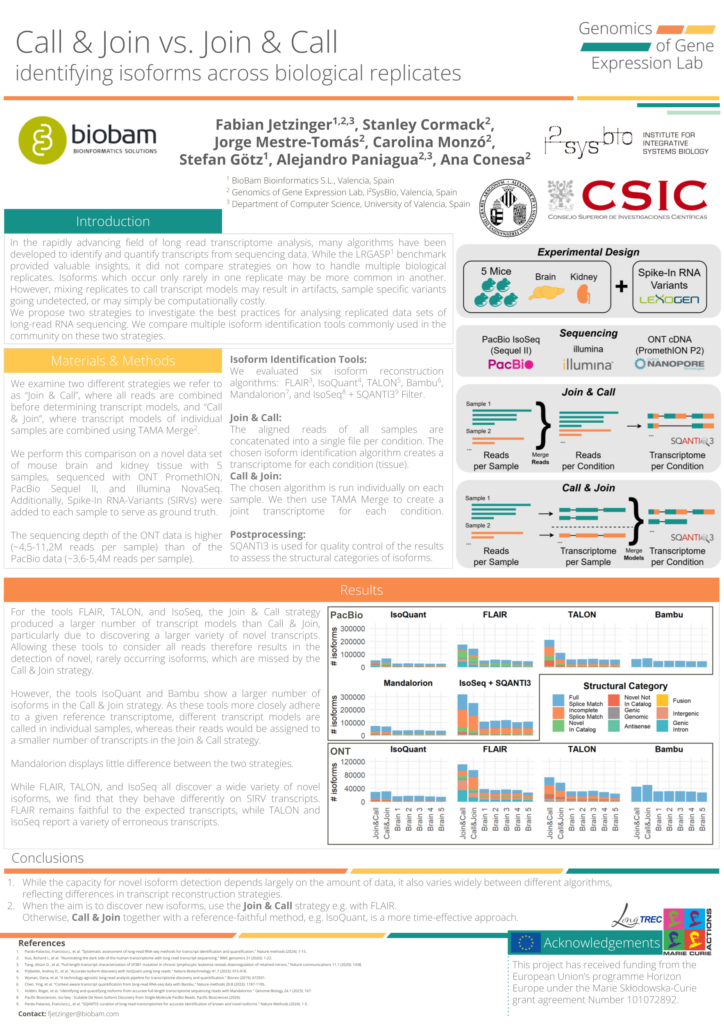
During this time, the LongTREC network also held a meeting to provide PhD students with in-depth training sessions from various experts, as well as attending the Long-Read Sequencing Uppsala 2024 conference. Fabian used this chance to present some of his recent work in the form of a poster: titled “Call & Join vs. Join & Call: identifying isoforms across biological replicates”, this ongoing research project conducted in collaboration with the ConesaLab aims to investigate strategies on how best to identify RNA isoforms from experiments with multiple samples. Many of the researchers visiting the conference, among them world-renowned experts, stopped by to discuss the preliminary results and expressed great interest in the work.
One of the most anticipated events of the conference was the LongTREC workshop on long-read transcriptomics, where network members provided an overview of their broad spectrum of research in this domain. They covered topics such as mapping, transcriptome reconstruction, metatranscriptomics, and single-cell technologies. Contributing to this workshop, Fabian gave attendees an overview of how OmicsBox can facilitate an end-to-end analysis of long-read RNA sequencing data.

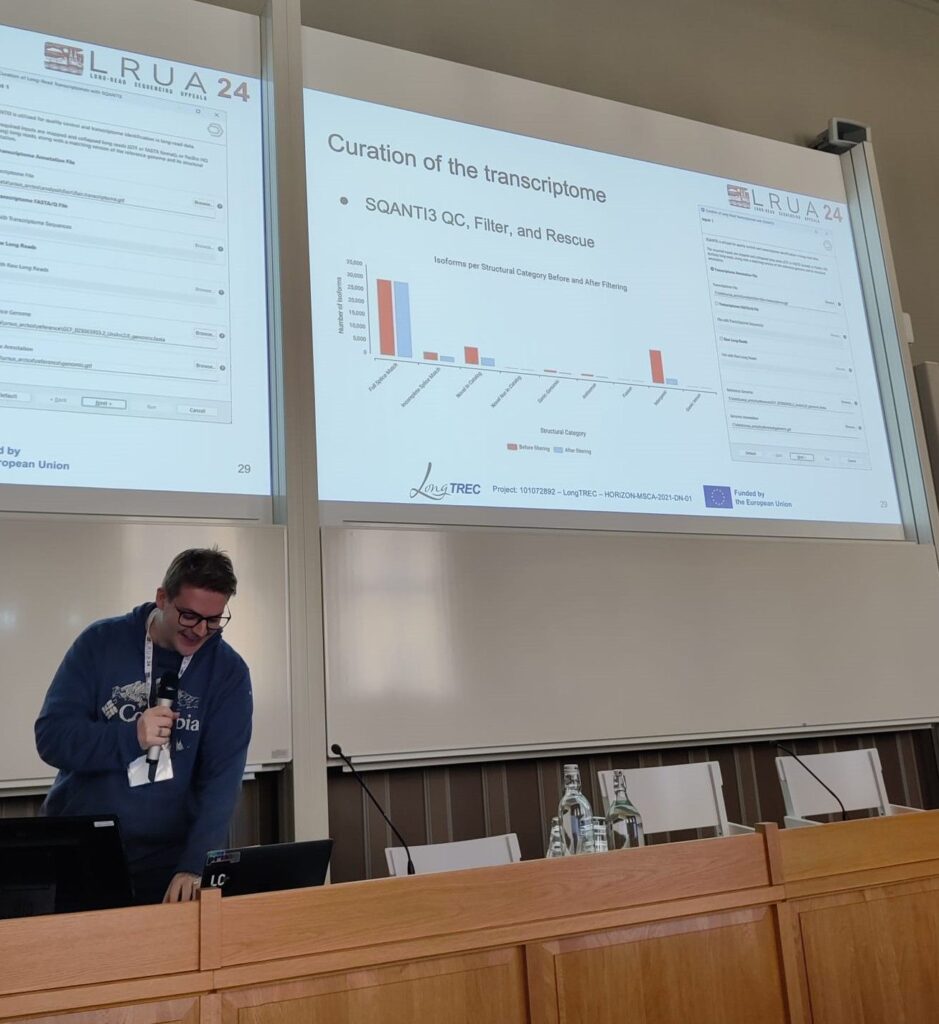
All in all, the stay in Sweden was a great success. Firstly, the direct collaboration with experienced developers of novel bioinformatics tools was a learning opportunity for both sides. This exchange of ideas and experiences between OmicsBox users, BioBam, and bioinformatics researchers lead to a variety of new ideas for improvements to research software. Secondly, the conference in Uppsala brought together experts from around the world. BioBam, which positions itself at the intersection of industrial applications and cutting-edge research, was a natural fit in this environment and had many productive exchanges with a wide variety of researchers in biology and bioinformatics.
Finally, we also want to thank our users who have provided helpful feedback on the use of the isONpipeline in OmicsBox. Thanks to our direct collaboration with the developers, future versions of OmicsBox will bring a wide range of further improvements to this tool, which has already proven itself to be invaluable to long-read transcriptomic research of non-model organisms.
About the Author




