Major Release Version 4
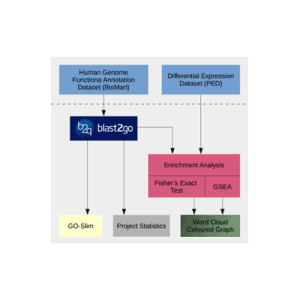
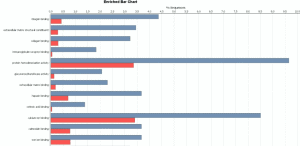
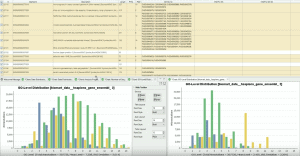
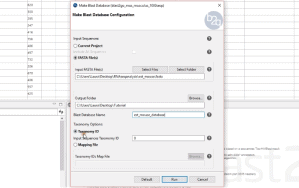
(Release Date: 13/09/2016) We are very happy to announce the release of Blast2GO 4. This new version contains many new bioinformatic features like Differential Expression Analysis, Gene Finding or Gene Set Enrichment Analysis. Blast2GO will update automatically if activated. Otherwise please download the latest version online from here. Feedback, questions, as well as feature requests, are most welcome. Please write