When running GeneFinding the sequences receive a name with the predicted genes.
The first part of the sequence identifier comes from the genome reference sequence name (de-novo assembly) and then a _orfx is appended, where x is a number.
Sometimes this name is not useful to proceed with downstream analysis or compare results from other experiments.
Is there any way in which I can attribute the 4,357 gene names to more standard gene IDs, such as rseq gene IDs or ENSG IDs?
The approach that can be followed is to replace the sequence name by the top hit from a reference (.fasta) retrieved by similarity.
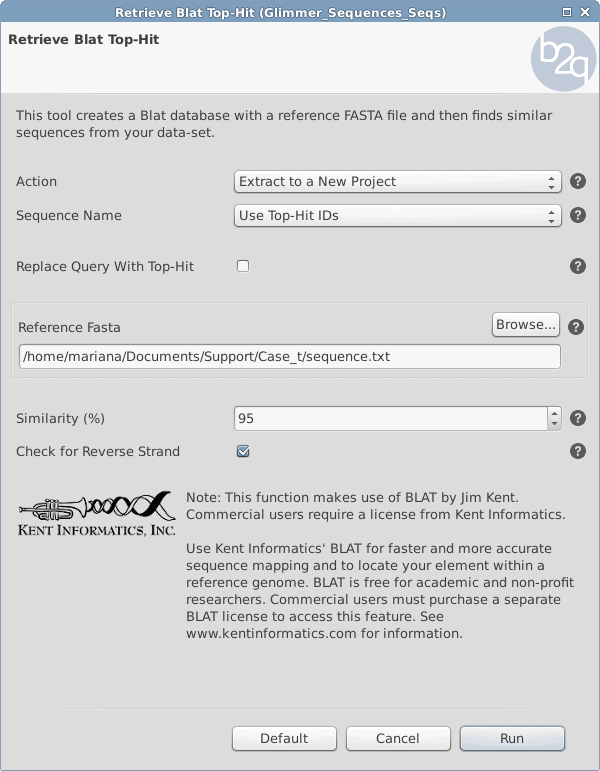
OmicsBox/Blast2GO offers the following feature under Tools > Retrieve Blat Top-hit which will search for similar sequences against a reference genome.
If the reference genome is available at the NCBI, this can be downloaded and then used to replace the names.
-
- Download reference genome (e.g. genes) from NCBI.
-
- Usually under Send to on the top right corner from the page e.g. Gene Features (Fasta Nucleotide).
-
- Under Tools > Retrieve Blat Top-hit choose the parameters like in Figure 1.
- Download reference genome (e.g. genes) from NCBI.
Figure 1: Retrieve Blat Top-Hit Parameters
The user will end up with a new project, where the sequences itself are from the gene finding project and the sequence names are the ones from the reference.
Note: The reference genome (genes) used in the feature can also be retrieved from BioMart from within OmicsBox/Blast2GO (see Load Sequences/ Annotation from a list of identifiers with Blast2GO) or from Load Fasta from Reference + GFF/GTF.