Sequence Analysis
High-Throughput Blast and InterProScan to perform fast sequence alignments and domain searches against reference datasets of your choice.
Gene Ontology Mapping
Link potential homologs and domains with available functional annotation from up-to-date and well-curated databases from UniProt and the Gene Ontology consortia.
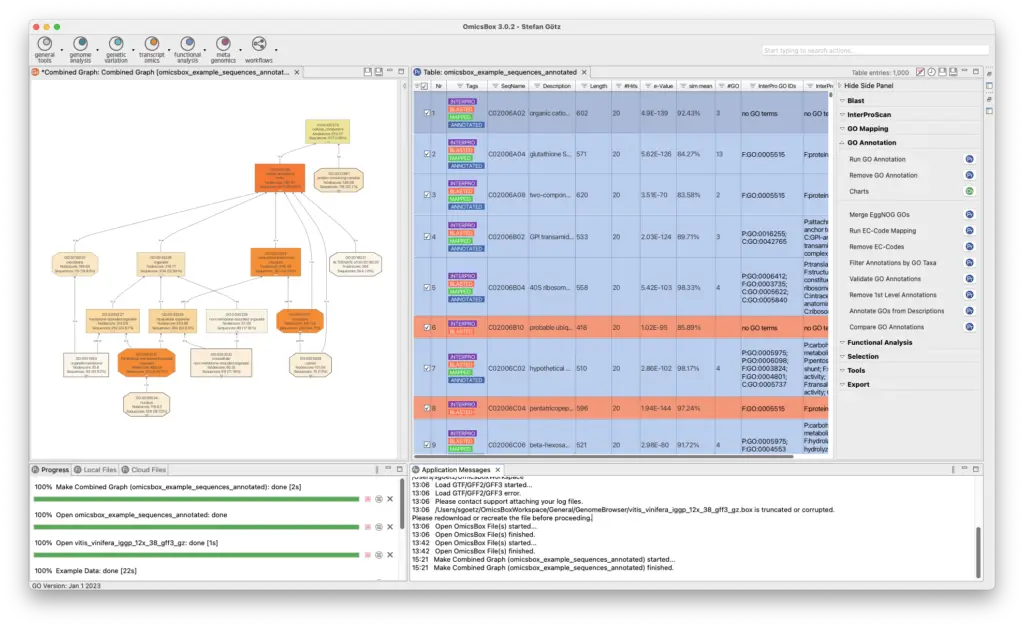
Blast2GO Annotation
The Blast2GO methodology allows assigning the most reliable functional labels to novel sequence datasets, combining different sources of annotations like protein domains, and orthology groups of basic sequence similarities, always taking into account annotation quality and gene ontology hierarchies.
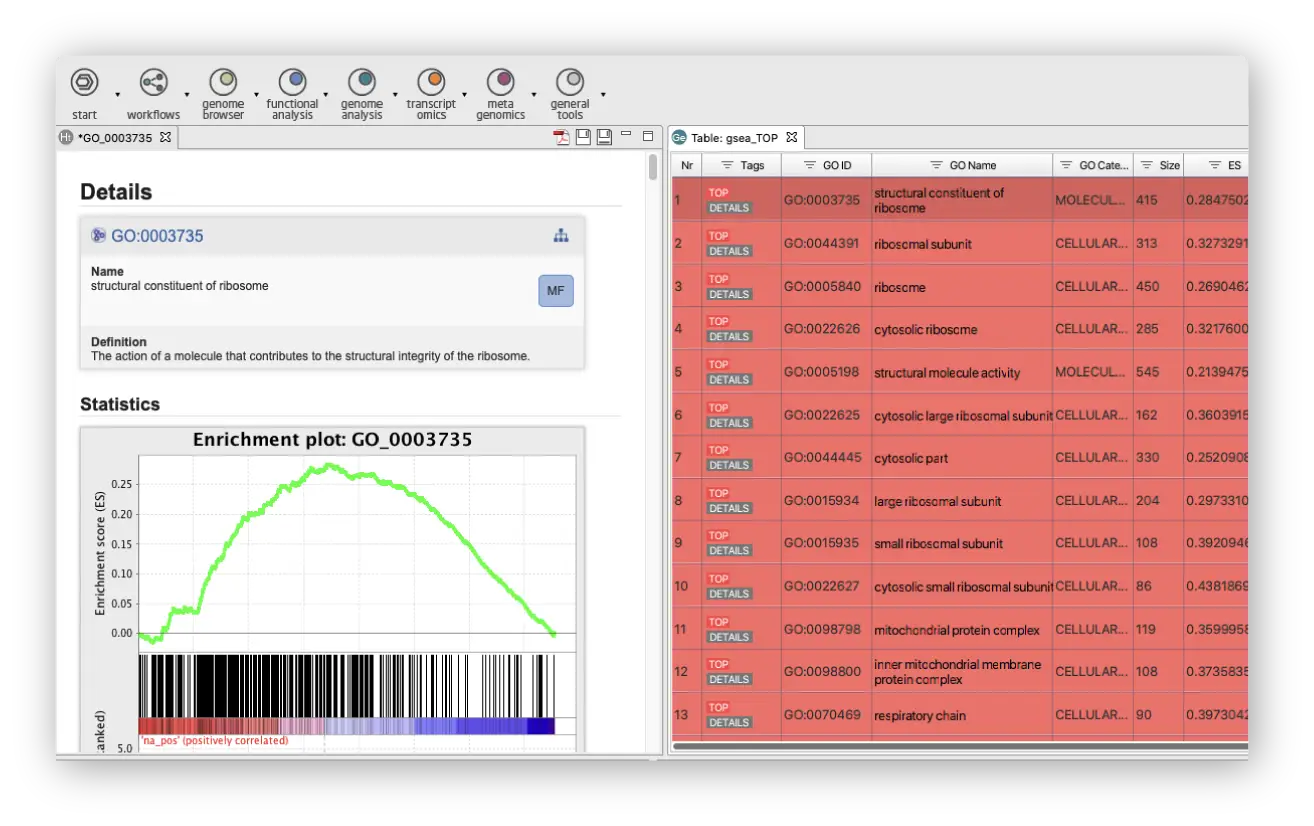
Enrichment Analysis
Use different enrichment analysis approaches like Fisher Exact Test and GSEA to identify over and under-represented biological functions.
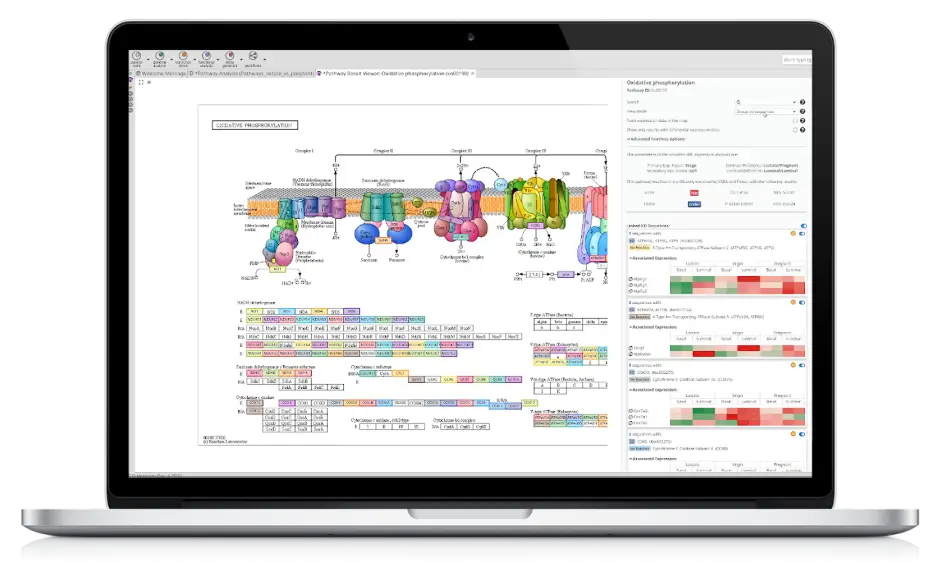
Combined Pathway Analysis
Identify Reactome and KEGG pathways for any set of sequences, use differential expression data to calculate pathway enrichment (GSEA) and a combined visualization to gain insights with ease.