BioBam Bioinformatics attended the 17th European Conference on Computational Biology ECCB in Athens, Greece. It is the main computational biology event in Europe and congregates scientists working in a variety of disciplines, including bioinformatics, computational biology, biotechnology, medicine, and systems biology.
We presented all the new features developed, during the last month, in Blast2GO:
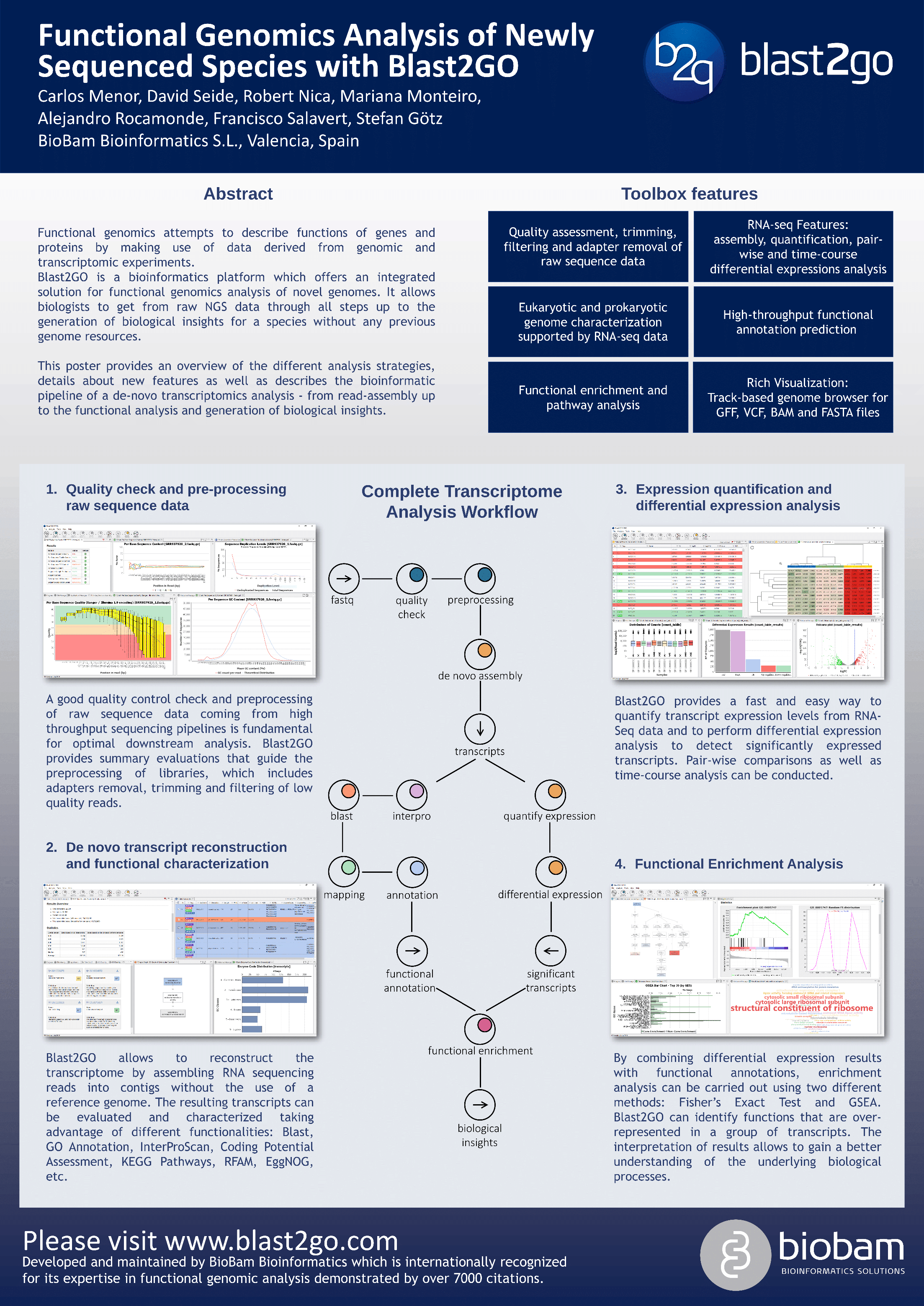
Functional Genomics Analysis of Newly Sequenced Genomes From Scratch with Blast2GO
Functional genomics attempts to describe the functions of genes and proteins by making use of data derived from genomic and transcriptomic experiments. The combination of omics data allows to better understand the relationship between genotype and phenotype on a system-wide scale.
Blast2GO is a bioinformatics platform which offers an integrated solution for functional genomics analysis of novel genomes. It offers biologists to get from raw NGS data through all steps up to the generation of biological insights for a species without any previous genome resources. The software runs out of the box, is biologist-oriented and has an intuitive design. The platform combines a rich graphical user interface with high-performance cloud computing. This allows high-throughput as well as exploratory analysis in just one place.
Most important features include:
-
- Structural characterizations with RNA-seq supported gene predictions
- RNA-seq features like assembly, quantification and pair- and time-course differential expressions analysis
- High-throughput functional annotation predictions
- Functional enrichment and pathway analysis
- Track-based genome browser for GFF, VCF, BAM and FASTA files
This poster provides an overview of the different analysis strategies, provides details about new features as well as describes the use-case of a non-model de-novo transcriptomics analysis covering the whole pipeline – from read-assembly up the functional analysis and generation of biological insights.