Introduction
You want: Perform functional annotation and analysis of its potential proteins.
You need: Predict all potential genes or coding regions before proceeding to the functional annotation: Gene-Finding
How can this be done?
-
- Use Glimmer, a set of algorithms which uses interpolated Markov models to distinguish coding from non-coding DNA in bacteria, archaea, and viruses. Glimmer has been developed at the Center for Computational Biology at Johns Hopkins University, Baltimore, USA which is also the home of tophat, bowtie and cufflinks among others popular bioinformatics tools.
- Use GeneMark, a family of gene prediction programs, which use species-specific inhomogeneous Markov chain models of protein-coding DNA sequence as well as homogeneous Markov chain models of non- coding DNA. GeneMark is developed at Georgia Institute of Technology, Atlanta, Georgia, USA.
- Use Prodigal. Prodigal, which name stands for Prokaryotic Dynamic Programming Genefinding Algorithm is a microbial (bacterial and archaeal) gene finding program developed at Oak Ridge National Laboratory and the University of Tennessee, USA. Prodigal is known to be a very fast gene recognition tool and a highly accurate gene finder which performs well also with high GC content genomes. Prodigal is based on log-likelihood functions and does not use Hidden or Interpolated Markov Models.
A brief review of these gene finding tools:
We describe here a basic review of 3 popular prokaryote gene prediction tools: Glimmer, GeneMark and Prodigal. We performed gene predictions for the Gram-positive bacterium Streptococcus thermophilus. (wikipedia)
We downloaded the complete genome (.fna) from NCBI and used Glimmer, GeneMark and Prodigal for gene prediction.Glimmer and Prodigal have been executed locally, by downloading the programs from their web pages. The exact steps and command used are provided at the end of this article. GeneMark has been executed online and results were obtained by email.
To test the performance in terms of recall and precision we performed a blastn of the predicted genes for each tool against the official genes published at the NCBI. The blast database has been created with the corresponding (.ffn) file. The blastn algorithm has been performed within Blast2GO PRO using LocalBlast.
The following table summarises the results of the three algorithms used to predict the genes of Streptococcus thermophilus.
In addition, the blastn results against the original data from NCBI, that contains 1914 genes, are also provided below.
| Glimmer | GeneMarkS | Prodigal | |
| # Predicted Genes | 1272 | 2019 | 1899 |
| # Hits (true pos) | 1252 | 1879 | 1832 |
| # No Hits (false pos) | 20 | 140 | 67 |
| Missing Genes (false neg) | 662 | 35 | 82 |
| Precision | 98.4% | 93.1% | 96.5% |
| Recall | 65.4% | 98.2% | 95.7% |
| #Seq < 100% sim | 3 | 17 | 10 |
The official gene prediction (NCBI) contains 1914 sequences. Based on the blastn results with 100% similarity, we recovered 1252 genes with Glimmer, 1879 with GeneMark and 1832 with Prodigal. While Glimmer obtains the highest precision it also shows the lowest recall in this test scenario. GeneMarkS has the best recall with 98.2%. However, the best overall performance has been obtained by Prodical. We believe that the results of all 3 tools could be improved by further fine-tuning of parameters, something we did not consider for this basic evaluation.
Continue to functional annotation in Blast2GO
The obtained fasta file containing the gene predictions can now be used in Blast2GO for the functional annotation. The standard steps here-fore would be blastx against bacteria, InterProScan, perform Gene Ontology mapping and the functional annotation step. The obtained information can now be used for further downstream analysis like the functional enrichment analysis of expression profiles (e.g. obtained via cuffdiff) and pathway analysis.
Popularity of Tools in terms of citations:
| Published Articles | Citations | Year |
| 943
1776 |
1997
1999 |
|
| 1296 698Total: 1994 |
1998 2001– |
|
| 1069 | 2010 |
Instructions to perform gene predictions with Glimmer, Prodigal and GeneMarkS:
First, we need to download Streptococcus thermophilus genome from NCBI via FTP or Entrez: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nucleotide&id=55821993&rettype=fasta
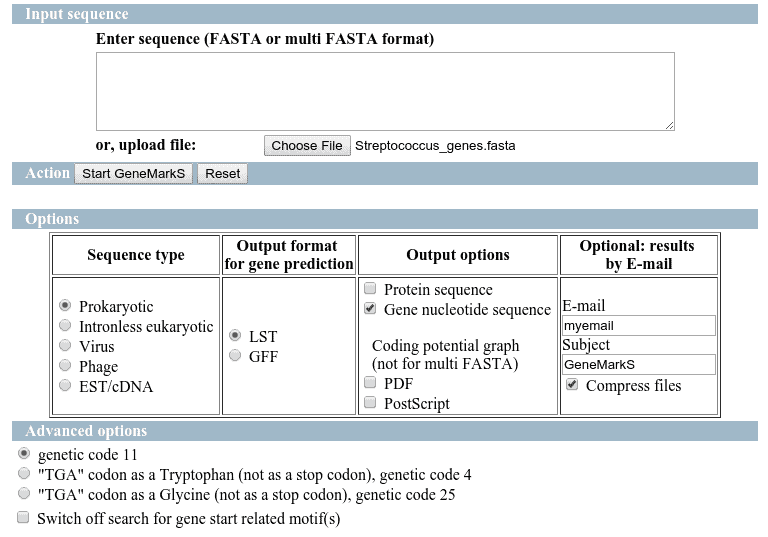
GeneMarkS
The predictions have been performed directly on GeneMarks webpage and the results have been retrieved on the email.
Glimmer
-
- Download Glimmer https://ccb.jhu.edu/software/glimmer/glimmer302b.tar.gz
- Extract Glimmer (see Glimmer notes for more information):
tar xzf glimmer302.tar.gz-
- Compile Glimmer
./src/make-
- Build Glimmer index for whole genome. Execute the following command from the bin folder.
./build-icm /path/to/index/output/filename/Prokaryota/Streptococcus/output.icm < /path/to/whole/genome/Prokaryota/Streptococcus/Streptococcus.fasta -
- Run Glimmer (percentages for ecoli start codons) – you will rectrieve 2 files .predict and .detail
./glimmer3 --start_codons atg,gtg,ttg --start_probs 0.83,0.14,0.03 --stop_codons tag,tga,taa --gene_len 110 --max_olap 50 /path/to/index/Prokaryota/Streptococcus/output.icm /path/to/output/filename/Prokaryota/Streptococcus/result/strep-
- Extract sequences from the .predict file
./extract -d -w /path/to/whole/genome/Prokaryota/Streptococcus/Streptococcus.fasta path/to/predict/filename/Prokaryota/Streptococcus/result.predict > path/to/output/filename/Prokaryota/Streptococcus/strep.fastaProdigal
-
- Download latest version of Prodigal https://github.com/hyattpd/prodigal/releases/
- Change the permissions of the prodigal.linux executable.
chmod 755 prodigal.linux-
- Run Prodigal:
./prodigal.linux -i /path/to/whole/genome/Prokaryota/Streptococcus/Streptococcus.fasta /path/to/output/filename/Prokaryota/Streptococcus/prodigal_predicted.fasta