Genome Analysis Module
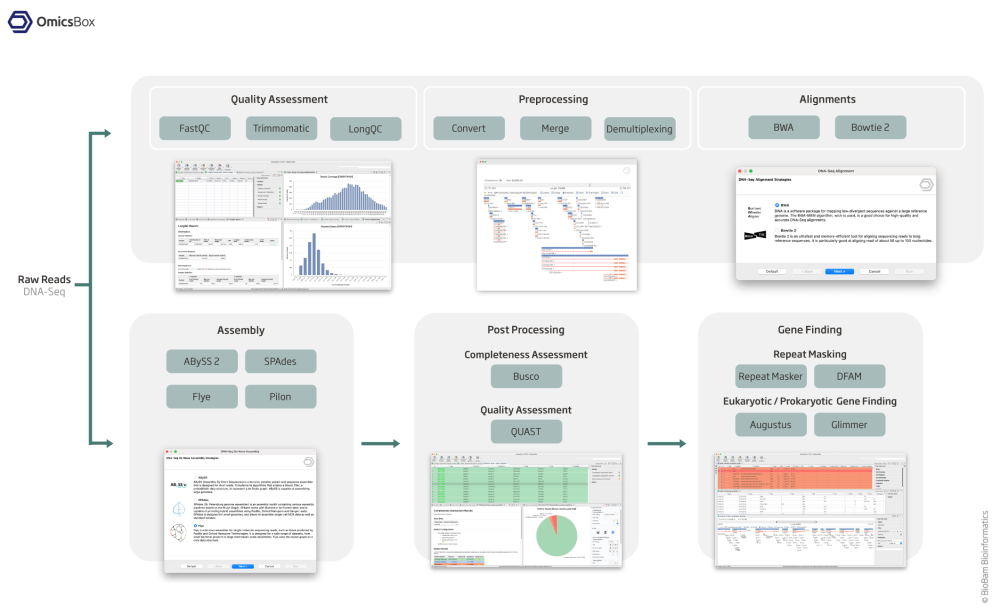
The OmicsBox Genome Analysis module allows to characterize and analyze newly sequenced genomes, from raw reads to gene structures in an efficient and user-friendly way.

Quality Control And Assessment
Use FastQC and Trimmomatic to perform the quality control of your samples, to filter reads and to remove low quality bases.
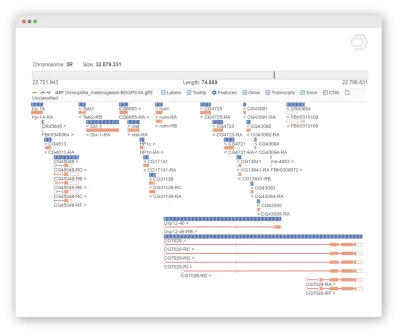
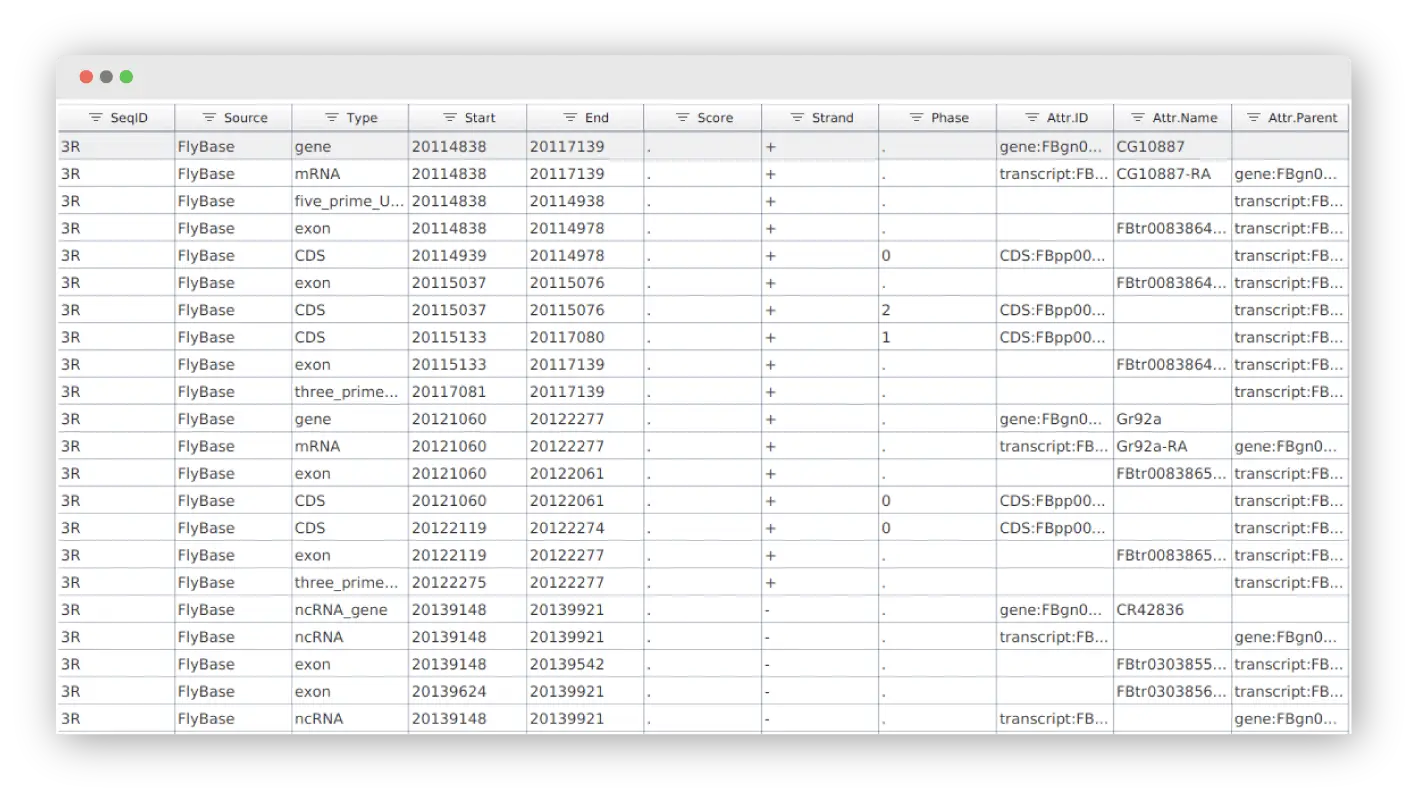
Genome Browser
Visualize your annotations in form of tracks to combine the genome sequences (.fasta) with alignments (.bam), intron-exon structure (.gff) and variant data (.vcf).
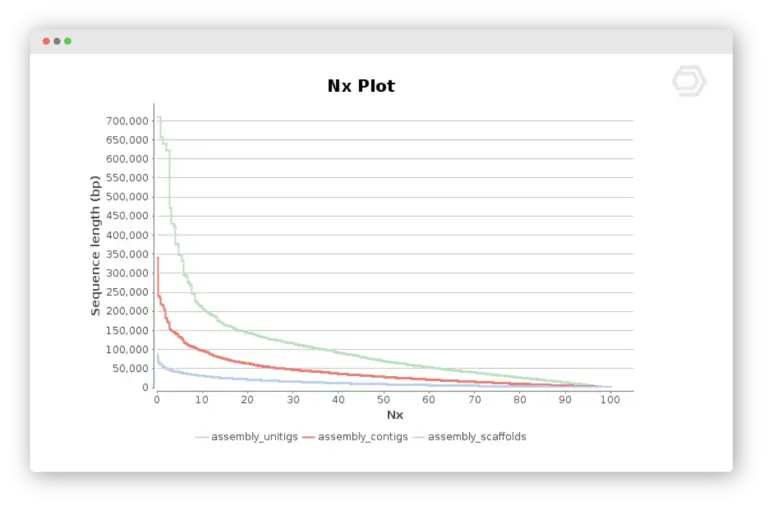
De-Novo Assembly
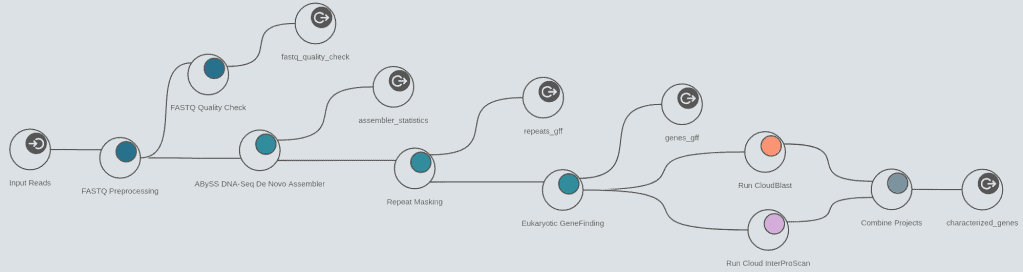
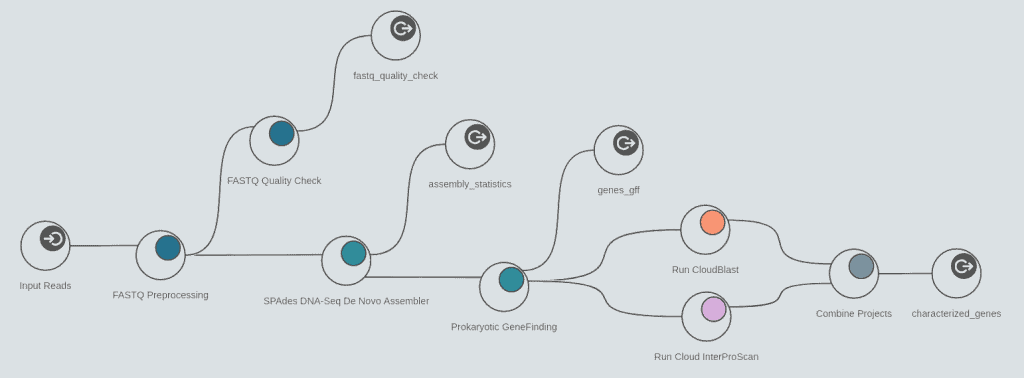
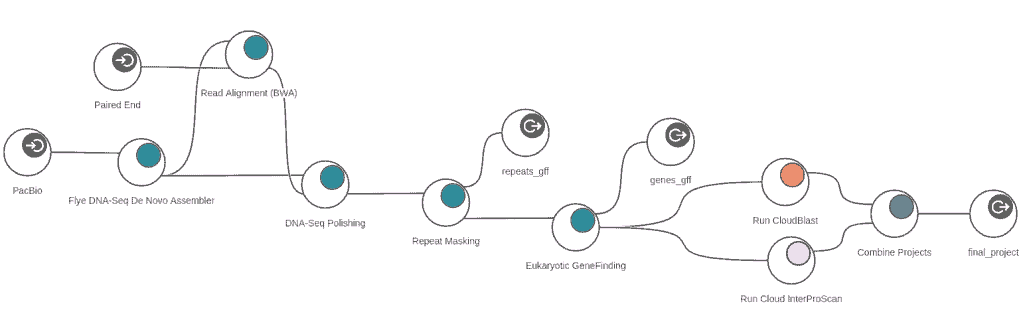
The assembly feature allows to reconstruct whole genome sequences without a reference genome or specific hardware requirements. Assemble sequencing data from both, short and long read technologies with 3 different algorithms: ABySS, SPAdes and Flye.
Repeat Masking and Gene Finding
Mask repeats of genome assemblies with RepeatMasker to improve downstream gene predictions.
Perform prokaryotic (Glimmer) and eukaryotic (Augustus) gene predictions to characterize genome structure.
Alignment and Polishing
Align short sequencing reads against large sequences with BWA or Bowtie 2, and correct draft assemblies from long reads with Pilon.
Multi-Locus Sequence Typing (MLST)
Characterize bacterial isolates unambiguously. This procedure considers the alleles present in (usually) seven well-characterized housekeeping genes.
- DNA-Seq de novo assembly with ABySS 2
- DNA-Seq de novo assembly with SPAdes
- DNA-Seq de novo assembly with Flye
- DNA-Seq alignment with BWA
- DNA-Seq polishing with Pilon
- Repeat masking with RepeatMasker
- Evidence-based eukaryotic gene predictions with Augustus
- Prokaryotic gene finding with Glimmer
- Genome Assembly Completeness Assessment with Busco