Once differential expression results are retrieved, it could be interesting to assign functional annotations to the differentially expressed genes and visualize the results in a single project. This is possible in OmicsBox.
Our starting point will be the transcriptomic sequences we used in the differential expression analysis. These sequences must be annotated (Functional Analysis > Blast, Blast2GO Mapping, and Blast2GO Annotation) and saved in an OmicsBox project.
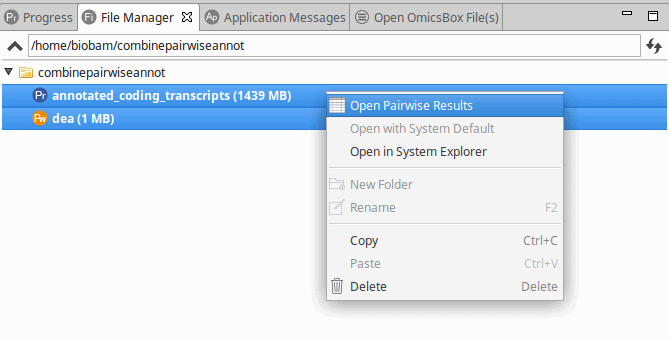
From here, it is possible to combine the differential expression results and the annotation via File Manager (View > File Manager). To combine the projects:
- Select both projects.
- Right-click on one of them and select “Open Pairwise Results“.
Figure 1: Combining Differential Expression Results and Functional Annotations via File Manager.
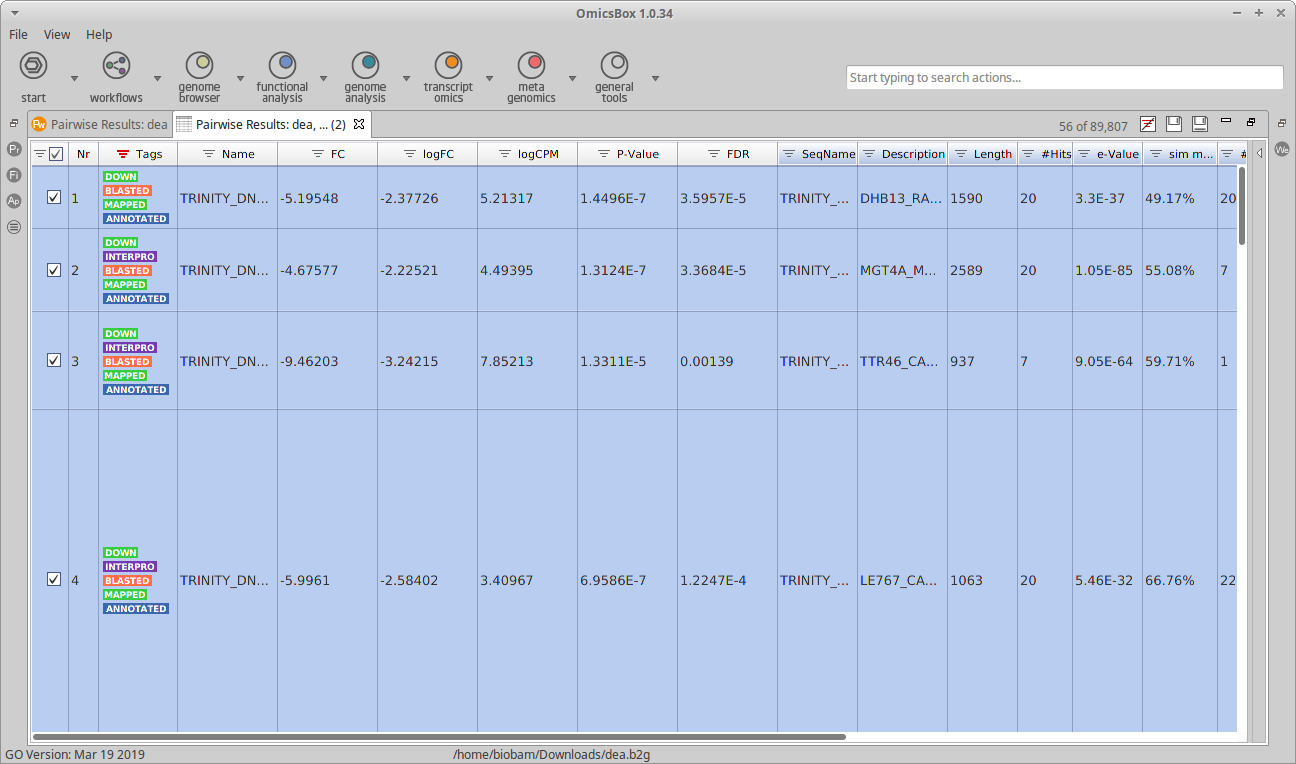
A new table should open now in OmicsBox, with the combined information of both projects. In this table, it is possible to explore the functional annotations of the up-regulated or the down-regulated genes using the Filters.
Figure 2: Table with Differential Expression and Functional Annotation data combined.
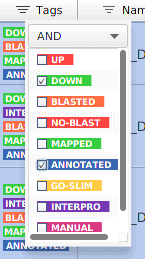
For example, imagine that we are interested in knowing the functional annotations of the down-regulated genes. We can use the filters in the Tag column: “DOWN” and “ANNOTATED”.
Figure 3: Filtering down-regulated and annotated genes.