It is possible to run Blast only on the differential expressed genes and not on all the data with OmicsBox.
One has to select only those sequences that have differential expressed genes in the OmicsBox project.
First, make sure that the name of the sequences in the project match the ones from the differential expression results.
Once confirmed, the starting point is the differential expression results, we will generate an ID List and then the sequences will be selected in the OmicsBox project.
Open the differential expression results and do the following:
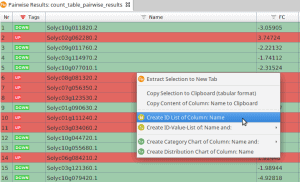
- Mark all rows using (Ctrl + A in windows and Linux or Cmd + A in Mac)
- Right-click on column Name.
- Choose Create Id-List of Column: Name (see Figure 1)
- A new tab will open.
- Save the ID List.
Figure 1: Create ID List of Column Name.
Now we will open the project with the sequences in order to only select the ones you want to run Blast.
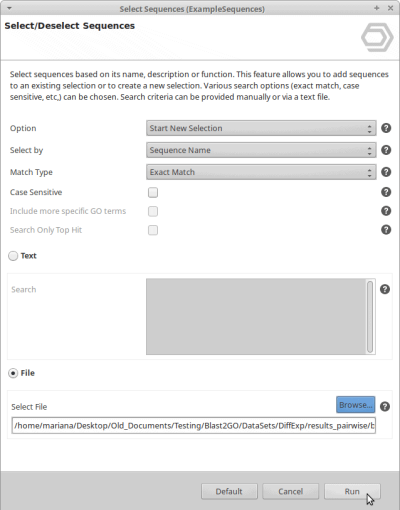
- Go to Functional Analysis > Tools (Select, Rename, Search) > Select > Select Sequences
- Now use Start New Selection, Sequence Name and Exact Match.
- Choose File and browse for the ID List that has been generated in the previous steps.
- Click Run
If the sequence matches, only those that have the exact name will be checked on the sequence project.
This means one can send only these sequences to Blast.