Using cloud resources efficiently is crucial for researchers dealing with large datasets. This article will show you how to use CloudBlast effectively, helping you save on computational costs and avoid common pitfalls.
OmicsBox allows you to perform NCBI Blast searches using a cloud system for high-performance computing. This system runs jobs in parallel and autoscales depending on demand. To control usage we use so-called CloudUnits. These units represent computation time (CPU seconds) on the AWS infrastructure. At the moment, only features related to sequence Alignments and Assembly consume units in OmicsBox, as they represent over 80% of the total cloud costs. For more information, please visit: Cloud Computation.
All OmicsBox subscriptions include a certain amount of units that can easily be recharged. However, to consume these resources wisely, you should take several aspects into consideration when performing Blast searches. Please remember that the consumption of units does not depend on the overall time it takes to Blast your dataset but on the amount of computational resources used.
When analyzing large sequence datasets for the first time, it’s important to proceed carefully to avoid unnecessary costs or delays. Please read the following recommendations to ensure efficient use of resources:
Consider Using DIAMOND
We recommend using the DIAMOND algorithm especially for large datasets. DIAMOND is optimized for handling large data volumes with reduced computational load, making it significantly faster while using fewer resources. This can help you save on CloudUnits while maintaining high sensitivity. DIAMOND is the default option in the Blast wizard, making it easy to get started with optimal settings. For more details see Buchfink B., Reuter K., & Drost H.G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nature Methods, 18(4), 366-368
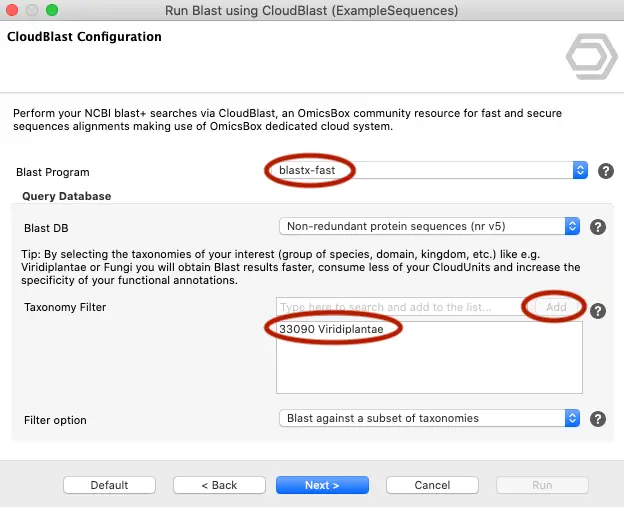
Reduce Your Search Space
Use a taxonomic filter. This saves time and computational resources (and units). Of course, this is just a recommendation, and the final decision depends on your research requirements and budget. Normally, if you are looking for potential homologous sequences (e.g., a plant species), you may want to consider only plant species and exclude the significantly large amount of bacterial genomes (>50%) in the NR protein database. This saves time and units.
Adjust Search Sensitivity
We recommend using the blastx-fast configuration, which basically increases the word-size of the alignments from 3 to 6. According to the NCBI, this should not alter your homology search results significantly but provide a performance increase. This configuration has been described more in detail by Shiryev et al. (2007). NCBI PubMed
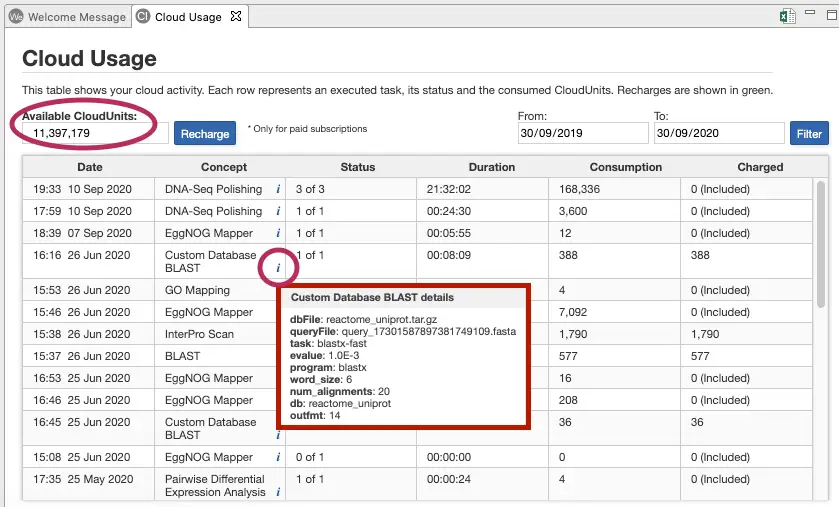
Check on your Cloud Usage
Use the “Cloud Usage” tab to review your unit consumption from within OmicsBox (from the “View” menu). Please see image below.
If you are new to CloudBlast and OmicsBox, we recommend trying your final blast parameters on a smaller dataset first. This avoids surprises. Use a small subset of your dataset (for example, 1000 sequences) to estimate unit consumption, then review and compare a few of your alignment results and the amount of consumed units.
Example
If you are blasting 1000 nucleotide sequences (contigs, CDS) against the NR database without a filter on a specific taxonomy group and with blastx (without the “fast” option), it means you are searching all 6 reading frames of all sequences against the world’s largest protein sequences collection with great sensitivity. Compared to a blast search against a plant subset with blastx-fast, this approach can reduce resource consumption by more than 10 times, translating into significant cost savings and improved time efficiency. The below numbers are from September 2020.
- 1000 nucleotide sequences with blastx-fast against the NCBI non-redundant database: 46 min. and 99173 CloudUnits
- 1000 nucleotide sequences with blastx-fast against the NCBI non-redundant viridiplantae subset: 26 min. and 7880 CloudUnits
Disclaimer
BioBam is committed to keeping cloud usage costs transparent and fair, with units serving solely to cover the actual cloud expenses. Units are pass-through items for the sole purpose of covering cloud expenses.