Sometimes databases provide the whole genome and the GFF or GTF files but not the exon or CDS FASTA files.
With OmicsBox/Blast2GO it is possible to load a Fasta sequences and to extract the exons or the CDS from the genome using the GFF file.
Use Case
For this example, the data used is from NCBI Bacteria Escherichia coli BW25113.
The sequences that will be loaded in Blast2GO will be the ones with feature exon (3rd column) in the GFF file and the given sequence name has to be chosen from the 9th column e.g. exon_id.
The GFF file looks the following:
Chromosome ena gene 190 255 . + . ID=gene:BW25113_0001;Name=thrL;biotype=protein_coding;description=thr operon leader peptide;gene_id=BW25113_0001;logic_name=ena
Chromosome ena mRNA 190 255 . + . ID=transcript:AIN30539;Parent=gene:BW25113_0001;Name=thrL-1;biotype=protein_coding;transcript_id=AIN30539
Chromosome ena exon 190 255 . + . Parent=transcript:AIN30539;Name=AIN30539-1;constitutive=1;ensembl_end_phase=0;ensembl_phase=0;exon_id=AIN30539-1;rank=1
Chromosome ena CDS 190 255 . + 0 ID=CDS:AIN30539;Parent=transcript:AIN30539;protein_id=AIN30539These are the steps to retrieve the exon sequences with exon id as sequence name
- Download the DNA (whole genome)
- Download GFF file
- In Blast2GO go to File > Load Sequences > Load Fasta from Reference + GFF/GTF
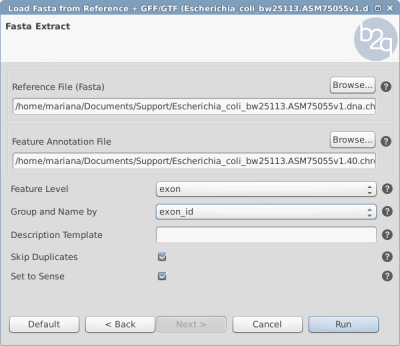
- See Figure 1 for parameters:
- Feature Level: exon
- Group and Name by: exon_id
- See Figure 1 for parameters:
Once loaded, a new project will be created in Blast2GO with the exon sequences and the SeqName corresponds to the exon_id, see Figure 2.
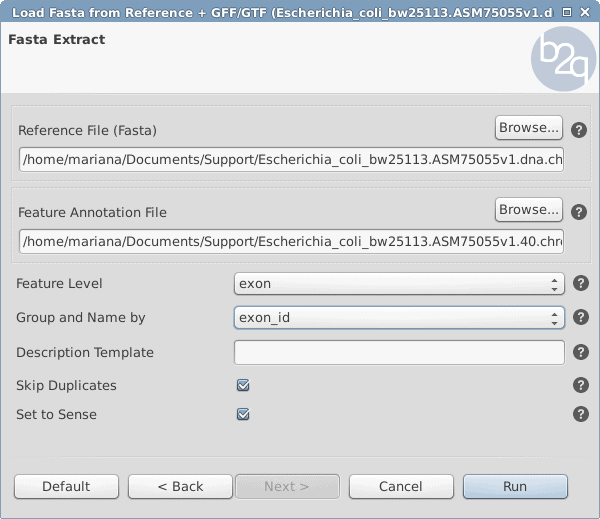 Figure 1: Load fasta sequences from the reference parameters window.
Figure 1: Load fasta sequences from the reference parameters window.
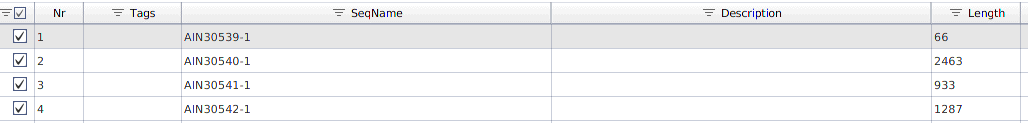 Figure 2: Exon sequences loaded in Blast2GO.
Figure 2: Exon sequences loaded in Blast2GO.
If you need more information about how to load FASTA sequences using a reference genome and a GFF file, please contact us.