OmicsBox allows running Blast locally. The blast algorithm will run on the user’s computer against a database that is installed locally.
In order to do so, we have to either download a pre-formatted NCBI database or format our own database (see this tutorial until step 3).
Pre-formatted databases can be downloaded directly from the NCBI ftp or via a Perl script provided by the NCBI.
Please follow the steps below to proceed with the download using the Perl script.
This tutorial will also provide information on how to create an nr subset database to run local Blast against it (Steps 5-7).
- Download the Blast+ executables (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/) from NCBI and extract it.
- The above-mentioned Perl script is located in the bin folder of the Blast+ executables and it can be used to download the desired pre-formatted database, e.g. nr.
To run the script, Perl has to be installed on the computer.perl update_blastdb.pl nr
- Once all .tar.gz files have been downloaded, they need to to be extracted. It is possible to do so in one go with a Linux command.
for file in *.tar.gz; do tar -xvzf $file; done
- The next 3 steps is to create the nr subset. If not interested continue to step 8.
- Search the Entrez Protein database and query for a taxonomic identifier e.g. “txid7742[ORGN]”
- Select “Send to File”, choose Format “GI list” and more recently “Accession List” and Create File
- Go to the folder where the uncompressed nr pre-formatted database is and run the following command.
GI list:/path/to/blast/binaries/blastdb_aliastool -gilist <gi_list_file> -db nr -out <out_db_name_of_your_choice>
Accession List:
/path/to/blast/binaries/blastdb_aliastool -seqidlist sequence_acclist.txt -db nr -out <out_db_name_of_your_choice>
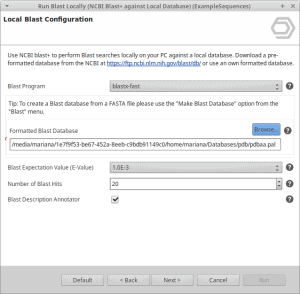
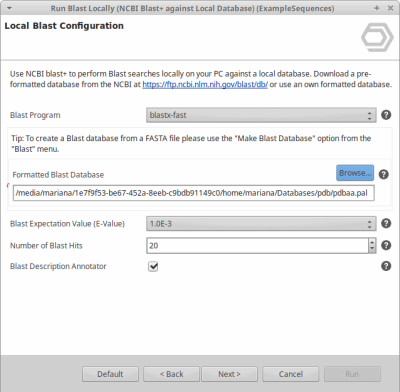
- That is all. You can now select the database file in the local blast dialog in OmicsBox.
Select the .pal file, if not present, change to “All Files” and select one of the extracted files.
In this example, we choose the pdbaa.pal file.