Input Reads
1
M
Predicted Genes
1
High-Quality Annotated
1
%
Processing Time
1
h
Use FastQC and Trimmomatic to perform the quality control of your samples, to filter reads and to remove low quality bases.
Identify Bacteria, Archaea, Fungi, Protozoa, and Virus down to strain level with Kraken 2. Gain insights from rich visualizations and test for the differential abundance of taxa.
Choose between MetaSPAdes and MEGAHIT to assemble large datasets easy and fast in the cloud.
Use FragGeneScan for plain reads and Prodigal for assembled data to identify and extract possible genes and proteins.
Obtain high-throughput functional annotations with EggNOG-Mapper and PfamScan. Results can be compared visually and differential abundance testing of functions is also available.
The Metagenomics Module of OmicsBox allows to combine and integrate all necessary steps for a complete microbiome data analysis in a flexible and intuitive way. Create custom pipelines for individual analysis strategies.
Take a look at this overview of metagenomic analysis options using OmicsBox.
The session covers quality control and assessment, taxonomic classification with Kraken2, and result interpretation. Also explores differential abundance analysis, illustrating how variations in microbial populations can be compared between samples.
Have a look at the poster we presented in Rotterdam at the Microbiome & Probiotics R&D & Business Collaboration Forum.
Get familiar with all new Modules and Features with a Free Trial or a Custom Demo.
OmicsBox works out of the box on any standard PC or laptop with Windows, Linux and Mac.
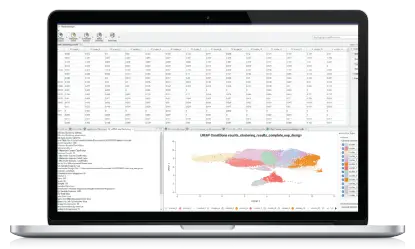
This example workflow shows a basic taxonomic classification of metagenomics data with OmicsBox. The reads are preprocessed (Trimmomatic) and a report is generated (FastQC). Kraken is used to identify and count all different operational taxonomic units (OTUs) for further interpretation. The spreadsheet-like result can be filtered and organized. The PDF report gives a clean overview of most abundant OTUs at different levels for each sample. Results also include intra- and inter-sample comparison charts. The whole workflow can be started within a few clicks and computationally intensive parts run transparently on the cloud.
Functional characterization of metagenomics data is a complex task. The OmicsBox Metagenomics module allows you to design streamlined workflows to easily combine the typically resource-demanding assembly step with gene predictions, as well as high-throughput functional annotation for large metagenomics data-sets. This example workflow shows the combination of MEGAHIT with Prodigal. Fast and comprehensive functional annotation is achieved with the integration of EggNOG-Mapper and PfamScan. Results are presented in form of spreadsheets and can be filtered and visualized with hierachical bar- and graph charts.
