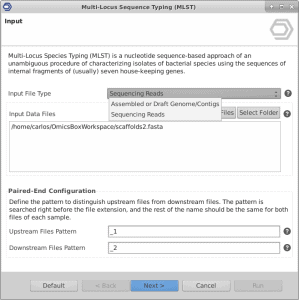
The Multilocus Sequence Typing Analysis Tool of OmicsBox (MLST) is a nucleotide sequence-based approach of characterizing isolates of bacterial species using the sequences of internal fragments of seven housekeeping genes.
This video shows step-by-step how to analyze bacterial sequences with the MLST App in OmicsBox. As input, users can provide either DNA-Seq reads in FASTQ or contigs/scaffolds in FASTA format. For each housekeeping gene, the different sequences present within a bacterial species are assigned as distinct alleles and, for each isolate, the alleles at each of the seven loci define the allelic profile or sequence type (ST).
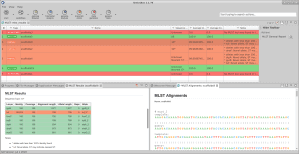
The MLST results are returned in tabular form. Sequence types assigned to each of the input samples (isolates) can be checked. In addition, each sample has a report containing details about the alignment against each housekeeping gene, and visualizations of those alignments that allow to find variations.
The MLST Analysis Tool can be installed from within OmicsBox. OmicsBox is available for Windows, Mac and Linux and can be downloaded here. A free trail key can be requested online here.
Find more details about MLST please read the online user manual or view the corresponding research citations below.
Some Screenshots:
References:
- Larsen MV et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. Journal of clinical microbiology, 50(4), 1355-61.
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K. and Madden TL. (2009). BLAST+: architecture and applications. BMC bioinformatics, 10, 421.
- Clausen PTLC., Aarestrup FM. and Lund O. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA. BMC bioinformatics, 19(1), 307.