Release OmicsBox version 1.2 (24th of October, 2019)
We are happy to announce the following updates for the genome analysis module.
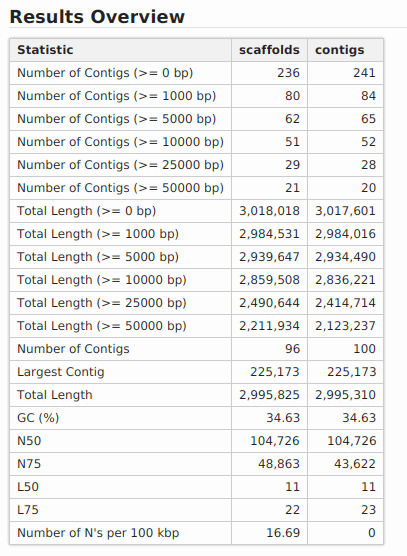
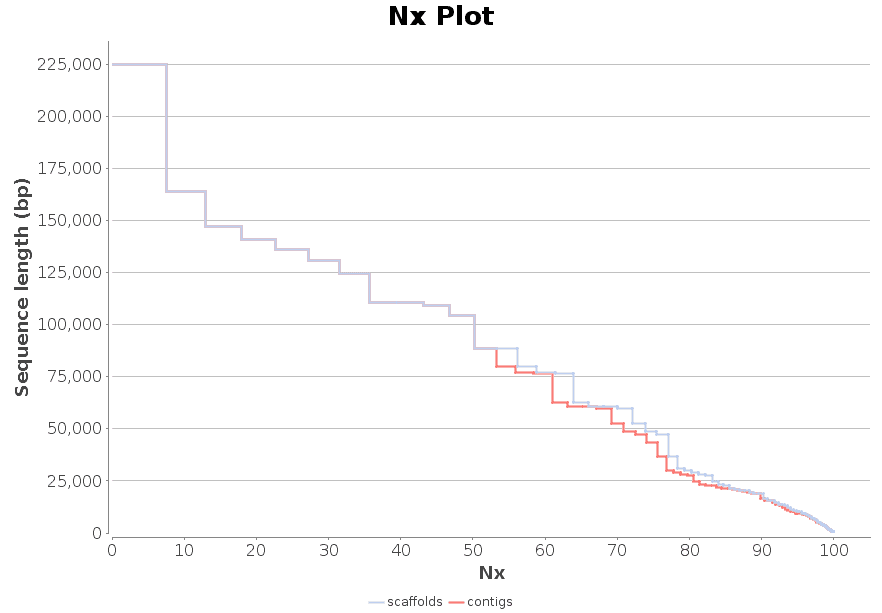
New feature includes a new DNA-Seq de novo assembly strategy based on SPAdes.
More details can be found below as well as in the online user manual and Genome Analysis Module website.
DNA-Seq de Novo Assembly: SPAdes
SPAdes (St Petersburg genome assembler) is an assembly toolkit containing various assembly pipelines based on the Bruijn Graph. SPAdes works with Illumina and IonTorrent data and is capable of providing hybrid assemblies using PacBio, Oxford Nanopore and Sanger reads. SPAdes is designed for small genomes and allows to assemble single-cell MDA data as well as standard isolates.
The SPAdes assembly pipeline consists of four stages:
- Assembly graph construction. SPAdes uses the multisized de Bruijn graph, implements new bulge/tip removal algorithms, detects and removes chimeric reads, aggregates biread information into distance histograms, and allows to backtrack the performed graph operations.
- k-bimer adjustment: SPAdes derives accurate distance estimates between k-mers in the genome using joint analysis of distance histograms and paths in the assembly graph.
- Constructs the paired assembly graph: Inspired by Paired de Bruijn graphs (PDBG) approach.
- Contig construction: SPAde constructs DNA sequences of contigs and the mapping of reads to contigs by backtracking graph simplifications.
Minor Improvements
- Repeat Masking improvements:
- Repeat Masking Database Update DFAM v.3
- Repeat Masking Compatibility with RepBase 2017 and 2018.
- DNA-Seq de Novo Assembly improvement:
- Improved wizard for sample selection.