
Mining Genes involved in Biodegradation of Tannery Wastes
This project shows how OmicsBox was used to functional annotate Moringa oleifera and to better understand the genome of the plant.

This project shows how OmicsBox was used to functional annotate Moringa oleifera and to better understand the genome of the plant.

This project shows how OmicsBox was used to functional annotate Moringa oleifera and to better understand the genome of the plant.

1. Introduction Juvenile rhesus macaques commonly suffer from idiopathic chronic diarrhea (ICD). This disease is characterized by inflammation of the colon and repeated bouts of diarrhea. It is a common cause of morbidity and mortality among macaques because it is unresponsive to drugs. The gut microbiome of macaques with ICD is characterized by abrupt changes when compared to healthy individuals.

We are happy to announce the latest version of OmicsBox 1.4, a very technical release of the OmicsBox base platform. We upgraded many internal libraries and updated to the latest version of many underlying software packages. These changes required a lot of code-refactoring and software testing to comply with all our internal security and quality standards. OmicsBox is now more
We are happy to announce that the OmicsBox Update Version 1.3 has now been released. OmicsBox provides now Long-Read support for DNA-Seq data analysis pipelines. Several new tools and updates improve the transcriptomics module and a new statistical package in the Metagenomics Module allow to identify functional differences among microbiome samples.
In this use case we will use the metagenomics tools included in OmicsBox to analyze the microbial communities of two different soda lakes from Brazil. The original study was carried out by Ana P. D. Andreote, et al., 2018 (doi: 10.3389/fmicb.2018.00244). Introduction Soda lakes are special ecosystems found across Africa, Europe, Asia, etc. These lakes show high levels of sodium
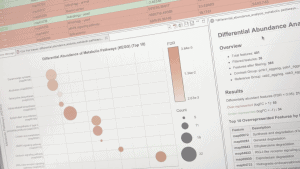
Any annotation of metagenomic sequences, whether it involves pathways, clusters of orthologous genes, species or any other taxonomic or functional classification can be compared across samples. These analyses are encompassed within the Differential Abundance Analysis, and they require robust statistical approaches to achieve reliable results because metagenomics datasets often show under-sampling and low signal-to-noise ratios. All differential abundance analyses can

In this use case we will use the metagenomics tools included in OmicsBox to analyze the microbial communities of two different soda lakes from Brazil. The original study was carried out by Ana P. D. Andreote, et al., 2018 (doi: 10.3389/fmicb.2018.00244). Introduction Soda lakes are special ecosystems found across Africa, Europe, Asia, etc. These lakes show high levels of sodium

The Multilocus Sequence Typing Analysis Tool of OmicsBox (MLST) is a nucleotide sequence-based approach of characterizing isolates of bacterial species using the sequences of internal fragments of seven housekeeping genes. This video shows step-by-step how to analyze bacterial sequences with the MLST App in OmicsBox. As input, users can provide either DNA-Seq reads in FASTQ or contigs/scaffolds in FASTA format.

